Tally 2.3
Tally-2.3 is a scoring tool based on a machine learning approach, which allows to validate the results of tandem repeat detection in protein sequences. As an input, Tally-2.3 uses tandem repeat region presented as a MSA of the repeats. An output lists Tally-2.3 and several other scores (Psim, entropy, p-value-phylo and parsimony) allowing the users to validate the quality of the examined tandem repeats.
N.B. Tally 2.3 is not a tandem repeat finder. If you want to detect tandem repeats in protein sequences, use Meta-Repeat-Finder or T-REKS.
For more details see:
Tally-2.0: upgraded validator of tandem repeat detection in protein sequences V. Perovic, J. Leclercq, N. Sumonja, F. Richard, N. Veljkovic and A. V. Kajava doi:10.1093/bioinformatics/btaa121
For details on our previous version see:
Tally: a scoring tool for boundary determination between repetitive and non-repetitive protein sequences. F. D. Richard, R. Alves and A. V. Kajava - Bioinformatics 2016, 1–7 doi:10.1093/bioinformatics/btw118
Tally-2.3 - Documentation
Tally-2.3 is a scoring tool based on a machine learning approach, which allows to validate the results of tandem repeat detection in protein sequences.
- 1. Input format
- 1.1 MSA only
- 1.2 FASTA
- 1.3 Fastally
- 2. Score
- Tally
- Psim
- p-value-phylo
- Entropy
- Parsimony
- 3. Example
- 3.1 Input
- 3.2 Output
1. Input format
1.1 MSA only
Insert MSA of repeats from one tandem repeat region like below :
Example :
1.2 FASTA
Insert MSA of repeats from one tandem repeat region in FASTA format.
Example :
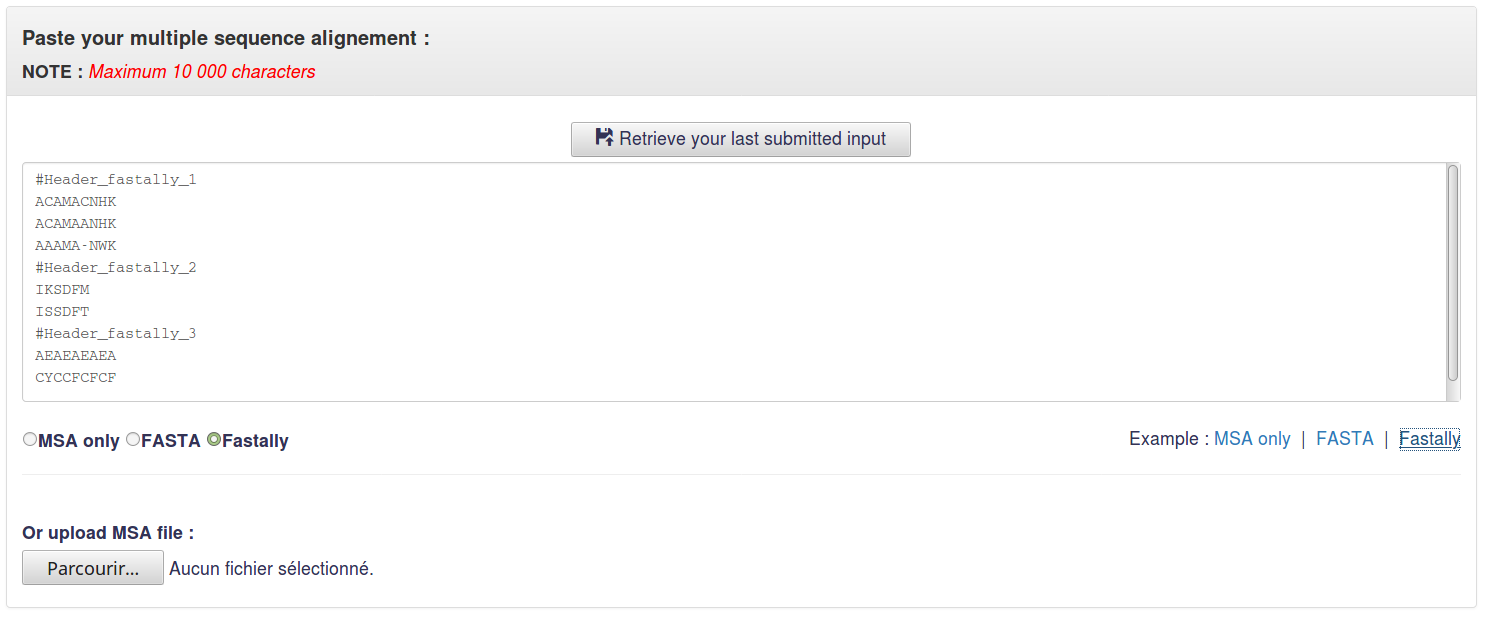
1.3 Fastally
Fastally format allows user to analyse several MSAs at one run.
In this format, headers started with # symbol separate MSAs of different tandem repeat regions.
Example :
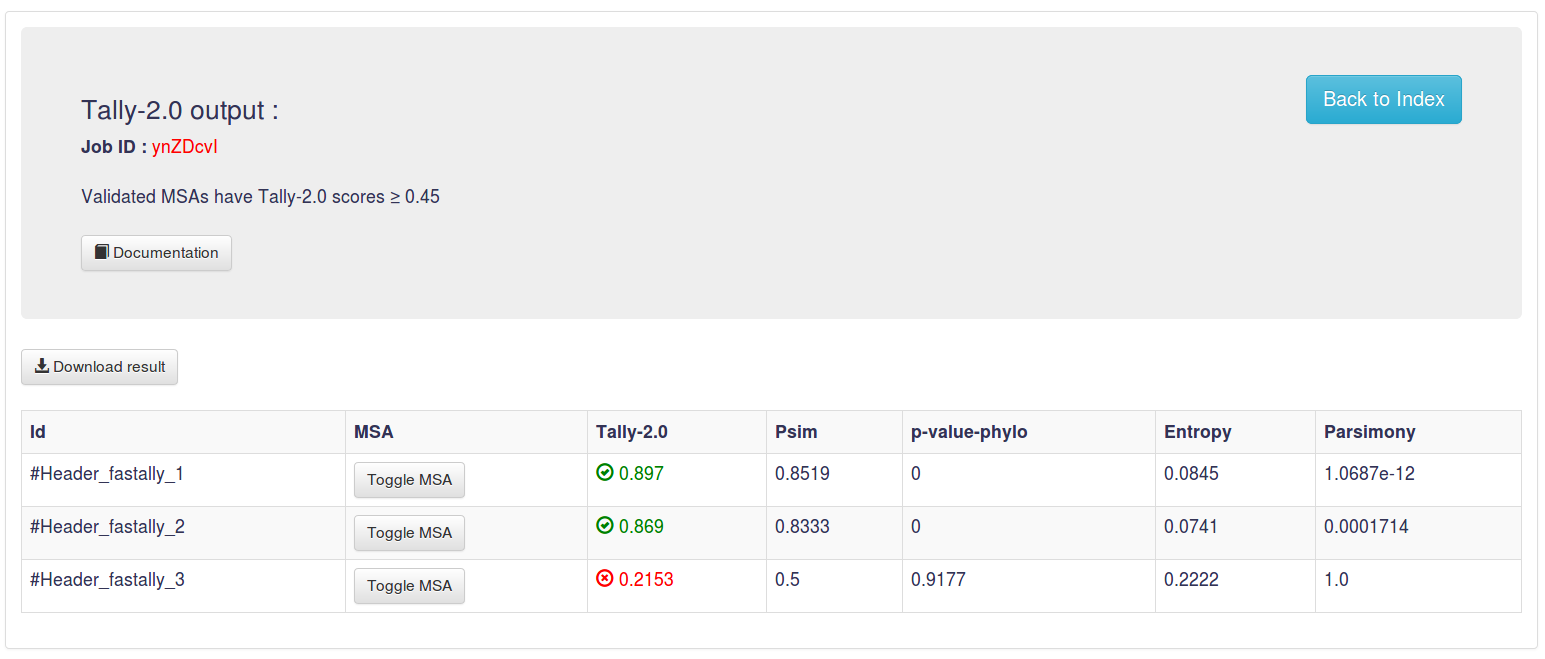
2. Score
Tally
Tally-2.3 score is obtained with machine learning approach. At a threshold of 0.5, established based on the maximization of F-score,
Tally-2.3 performs at a level of 89% sensitivity, while achieving a high specificity of 89% and an Area Under the Receiver Operating Characteristic Curve of 96%.
Validated MSAs have Tally-2.3 scores ≥ 0.5.
Psim
Psim is a score relying on the Hamming distance between the repeats and their consensus sequence.
Validated MSAs have Psim ≥ 0.7. [Psim documentation]
p-value-phylo
Validated MSAs have p-value-phylo scores ≤ 0.001. [Schaper et al.,2012]
Entropy
See : Entropy score definition
Parsimony
See : Parsimony score definition
3. Example
3.1 Input
3.2 Output