HELP
Input page
This is where you must provide information about your protein structure by entering the 4 letter PDB ID and its chain ID. A corresponding file in PDB format is downloaded automatically from PDB web site (www.rcsb.org).Output page
Output has three panels
- At the bottom, a panel of the overall schematic representation of protein sequence is shown, with the location of detected tandem repeats (alternating red and blue boxes). By default, the result of the detection module that yields the best results (in accordance with Q- and R- scores, see Annex 1 below) are shown. By clicking on “Show all TRs” one can see the results of all detection modules. To visualize two other panels, it is necessary to click on the blue link with the name of detection module.
- Viewer of the 3D structure. Repetitive elements are colored by alternated red and blue colors.
- Viewer of multiple sequence alignment of repetitive elements. It is based on the structural superposition of the repeats. Regions of the secondary structures are underlined (alpha- helices are in black, beta-strands are in red). Amino acids that are buried in the 3D structure are outlined by dark grey background. Color of amino acids depends on their physico- chemical properties.
FAQs?
- In your URL bar, enter: chrome://flags/#enable-npapi
- Click the Enable link for the Enable NPAPI configuration option.
- Click the Relaunch button that now appears at the bottom of the configuration page.
ANNEX I
Ranking Tandem Repeat (TR) candidates by R-score and Q-score
TR candidates found by different detection modules can overlap. In the final output, it is instrumental to identify these overlapping TRs and to choose among them the best TR representatives. Therefore, we cluster the TRs by using the Average Linkage Clustering method with the distances between two TRs equal to proportion of non-overlapping region of the shorter TR. A chosen distance cutoff retains within the clusters only those that were overlapped by more than 50%. Among the overlapping TRs of a cluster, we give a preference to the TRs, which cover the largest portion of the protein and have the smallest repeat unit. For this purpose, we have developed the ranking score (R-score):
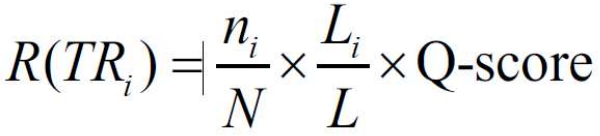
where L is the length of the sequence, which contains all overlapping TRs from the cluster. N is the maximal number of repeat units in one TR candidate in the cluster. ni and Li are the number of repeat units and total length of the ith TR, respectively. The Q-score evaluates the quality of a repeat Multiple Structural Alignment of a TR-candidate. The repeats of the TR candidate are aligned in Multiple Structural Alignment by using 3DCOMB program (Wang et al., 2011). In this Multiple Structural Alignment, an indel is considered as an additional symbol.
Based on the obtained Multiple Structural Alignment, values of Multiple Conformational Alphabet Alignment (MCAA) and Multiple Residue Contact Alignment (MRCA) are derived (see Appendix 6-7 in Phuong DO VIET thesis). MCAA uses the CA-1 conformation alphabet and MRCA uses information about residue contacts between the repeats. The Q-score of the Multiple Structural Alignment is defined as a linear combination of TM*-score (the maximal TM-score among the TM-scores of each pair of repeats), MRCA and MCAA values. Based on the ranking TAPO outputs five best TR candidates from each cluster.