General information:
The Cross-Beta DB website facilitates efficient exploration of the database. Users can
query and select amyloid regions (AR) of interest, view detailed information, and download
data in JSON or CSV formats. Since entries with identical protein sequences are grouped
together, some may have multiple associated structures.
Querying the database:
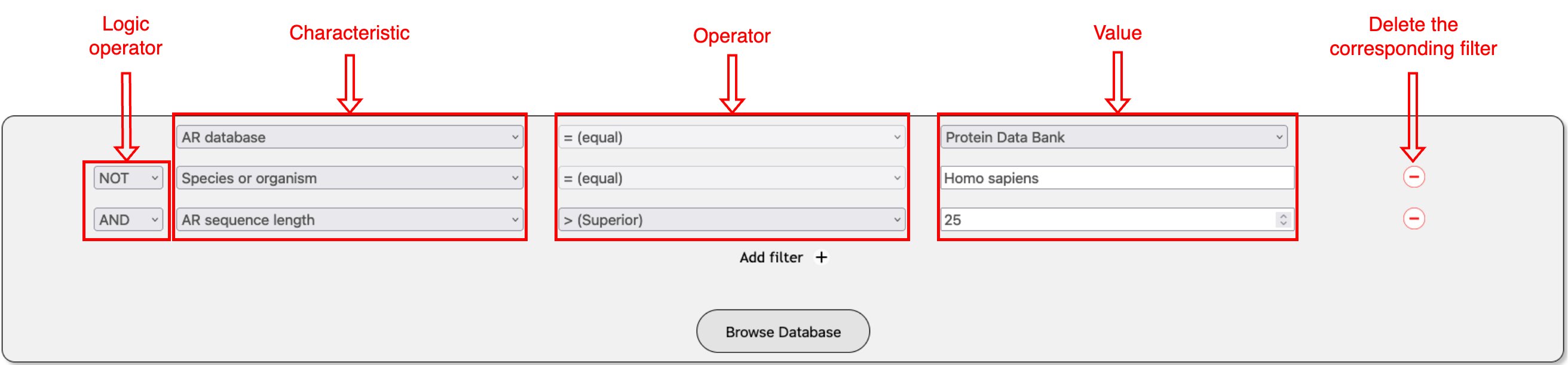
Use the Search section on the Home page to formulate queries by customizing
various elements. Queries consist of filters, which can be added using "Add filter (+)"
or removed by clicking the red minus sign (-).
Each filter includes three components: Characteristic (left), Operator (middle), and
Value (right). You can also specify the logic operator for each filter as "And" or "Not."
Once your query is built, click the "Browse Database" button to view the results.
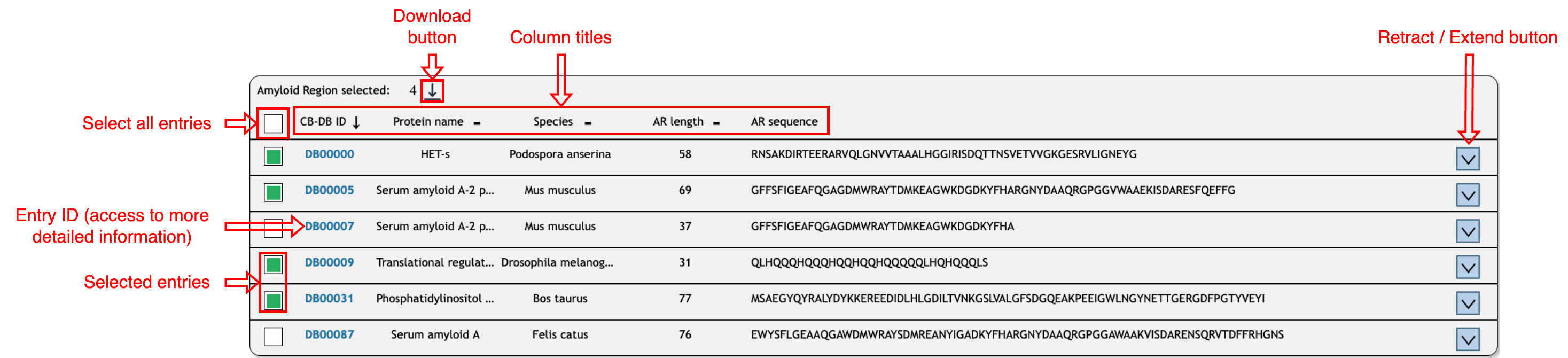
Browse and download query result:
The query results are displayed below, providing general information such as protein
name, species, amyloid region length, and sequence. Results can be sorted by "CB-DB ID,"
"Protein Name," "Species," or "Length" by clicking on the respective column header.
To download results, select entries by clicking the corresponding checkboxes or select
all by clicking the checkbox near the column title. Then, click the download icon and
choose a file format (.CSV, .JSON, or .FASTA). Note: Some values, such as author lists,
are comma-separated. To display CSV files correctly, set the column separator to ";".
If the sequence, protein name, or species name is too long to display fully, use the
"Retract/Extend" button to view the complete information. For additional details about
an entry, click its ID.